GitHub - tgac-vumc/ACE: Absolute Copy Number Estimation using low-coverage whole genome sequencing data
Absolute Copy Number Estimation using low-coverage whole genome sequencing data - tgac-vumc/ACE
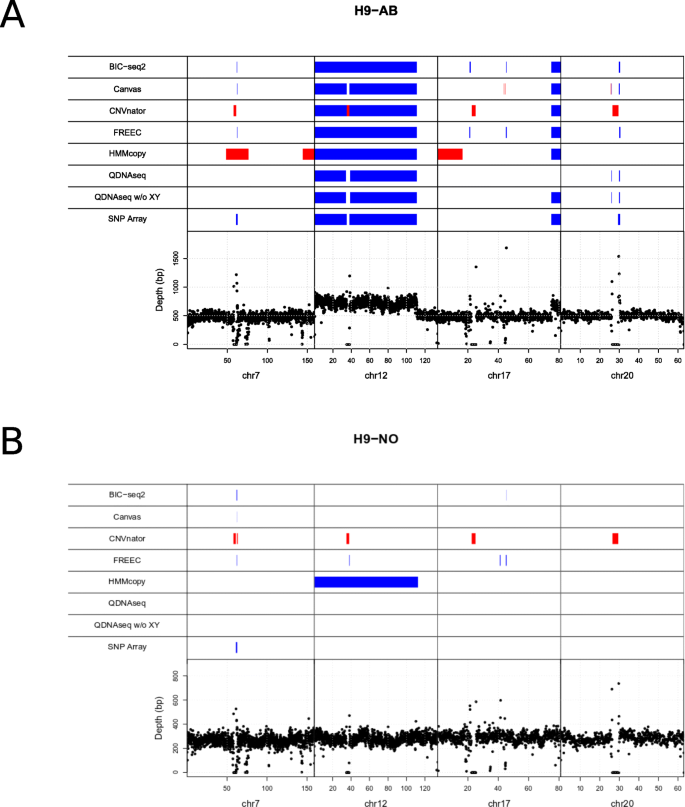
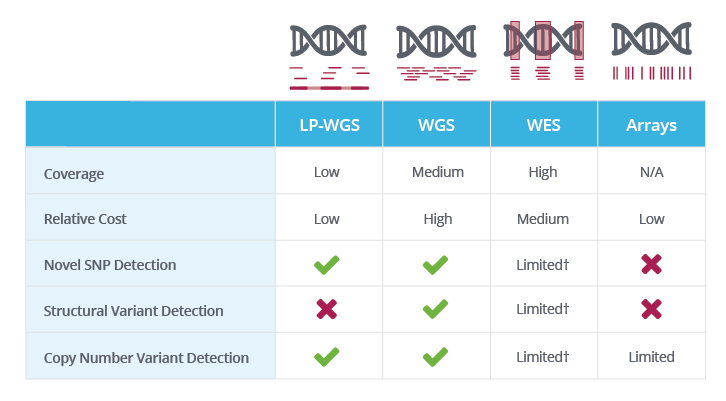
Evaluation of tools for identifying large copy number variations from ultra- low-coverage whole-genome sequencing data, BMC Genomics

Detection of Copy Number Variation using Shallow Whole Genome Sequencing Data to replace Array-Comparative Genomic Hybridization Analysis
Bamgineer: Introduction of simulated allele-specific copy number variants into exome and targeted sequence data sets

GitHub - Nealelab/whole_genome_analysis_pipeline

American Society for Clinical Pharmacology and Therapeutics - 2019 - Clinical Pharmacology & Therapeutics - Wiley Online Library
QIAGEN Omicsoft Copy Number Variation Analysis tutorial

Absolute copy number fitting from shallow whole genome sequencing data
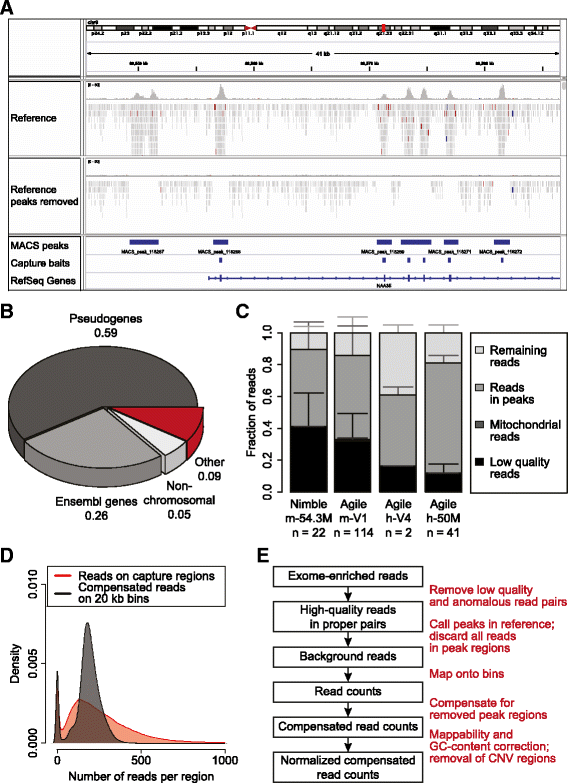
CopywriteR: DNA copy number detection from off-target sequence data, Genome Biology

Accucopy: accurate and fast inference of allele-specific copy number alterations from low-coverage low-purity tumor sequencing data, BMC Bioinformatics