Fitting to a Cryo-EM Map using MD Simulation
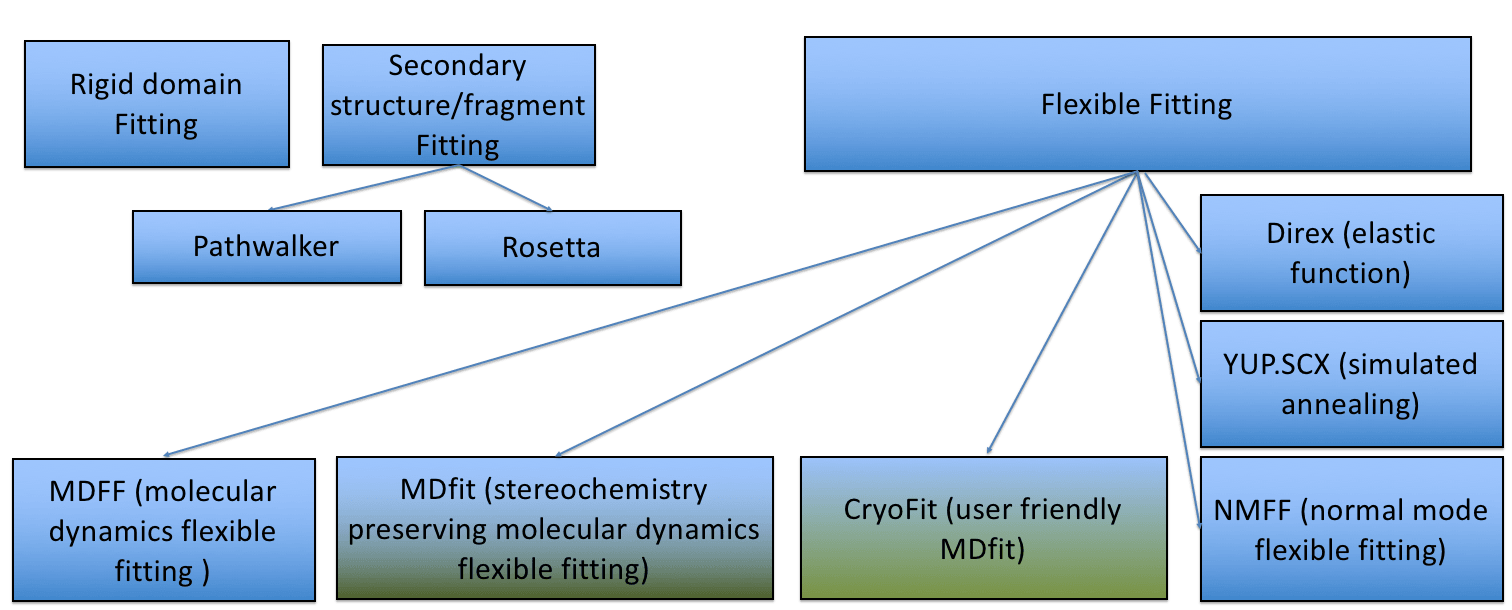

Atomic structure models are refined into a cryo-EM density using

MD-Fitting a Membrane Protein - Molecular Biophysics Stockholm
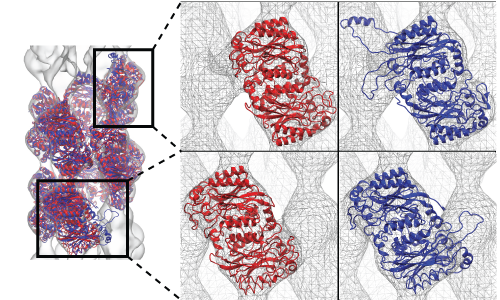
Molecular Dynamics Flexible Fitting
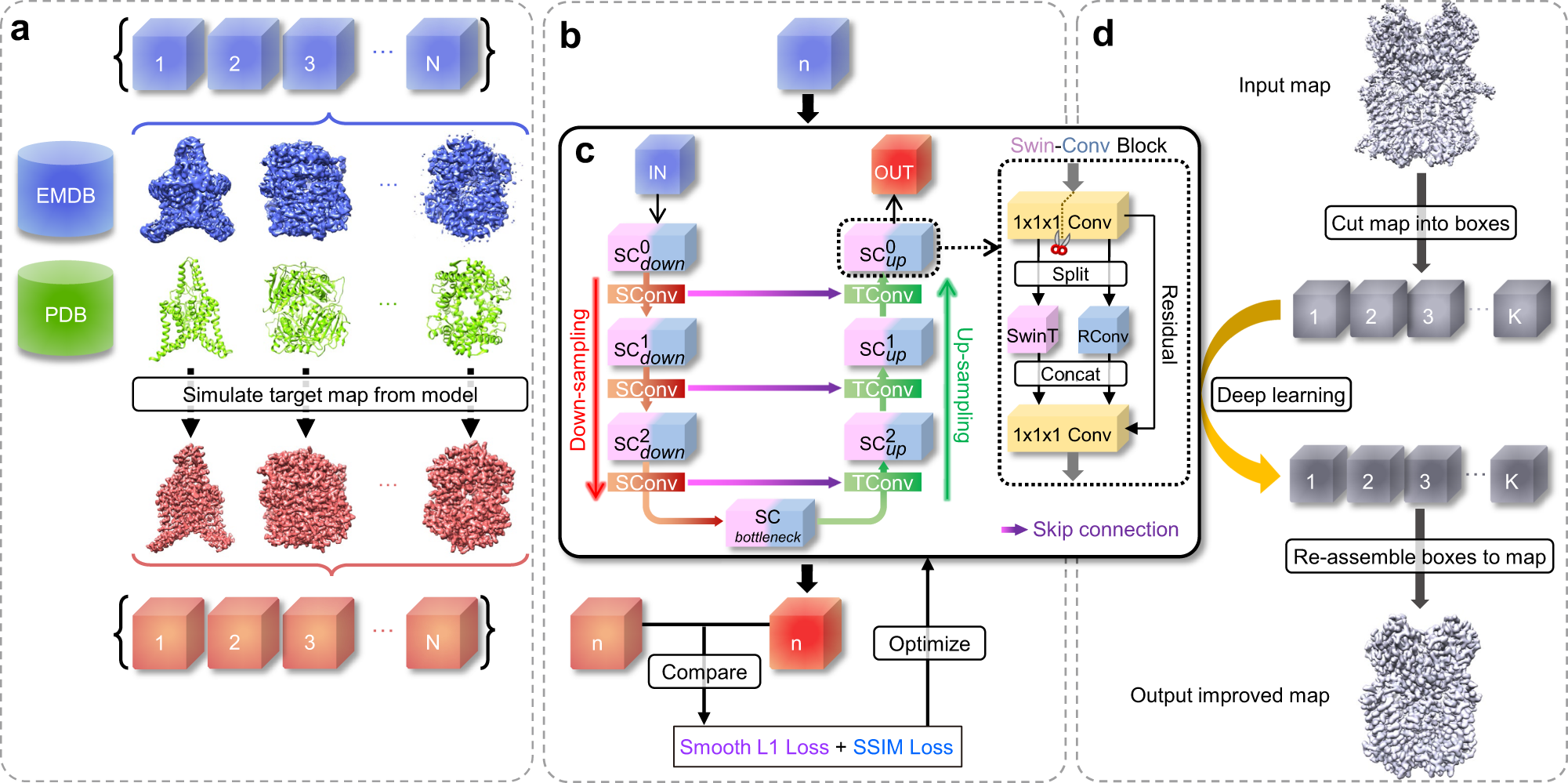
Improvement of cryo-EM maps by simultaneous local and non-local deep learning
Molecular Dynamics Flexible Fitting
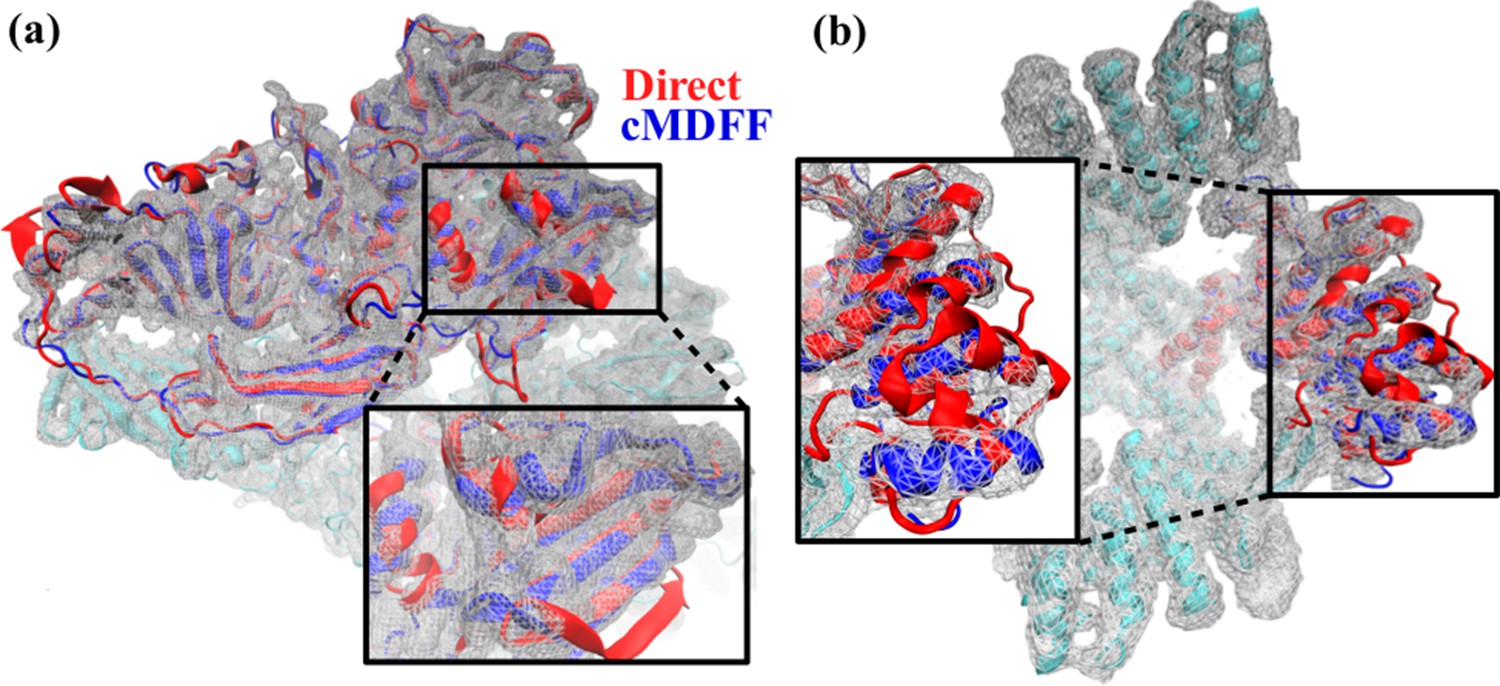
Molecular dynamics-based refinement and validation for sub-5 Å cryo-electron microscopy maps
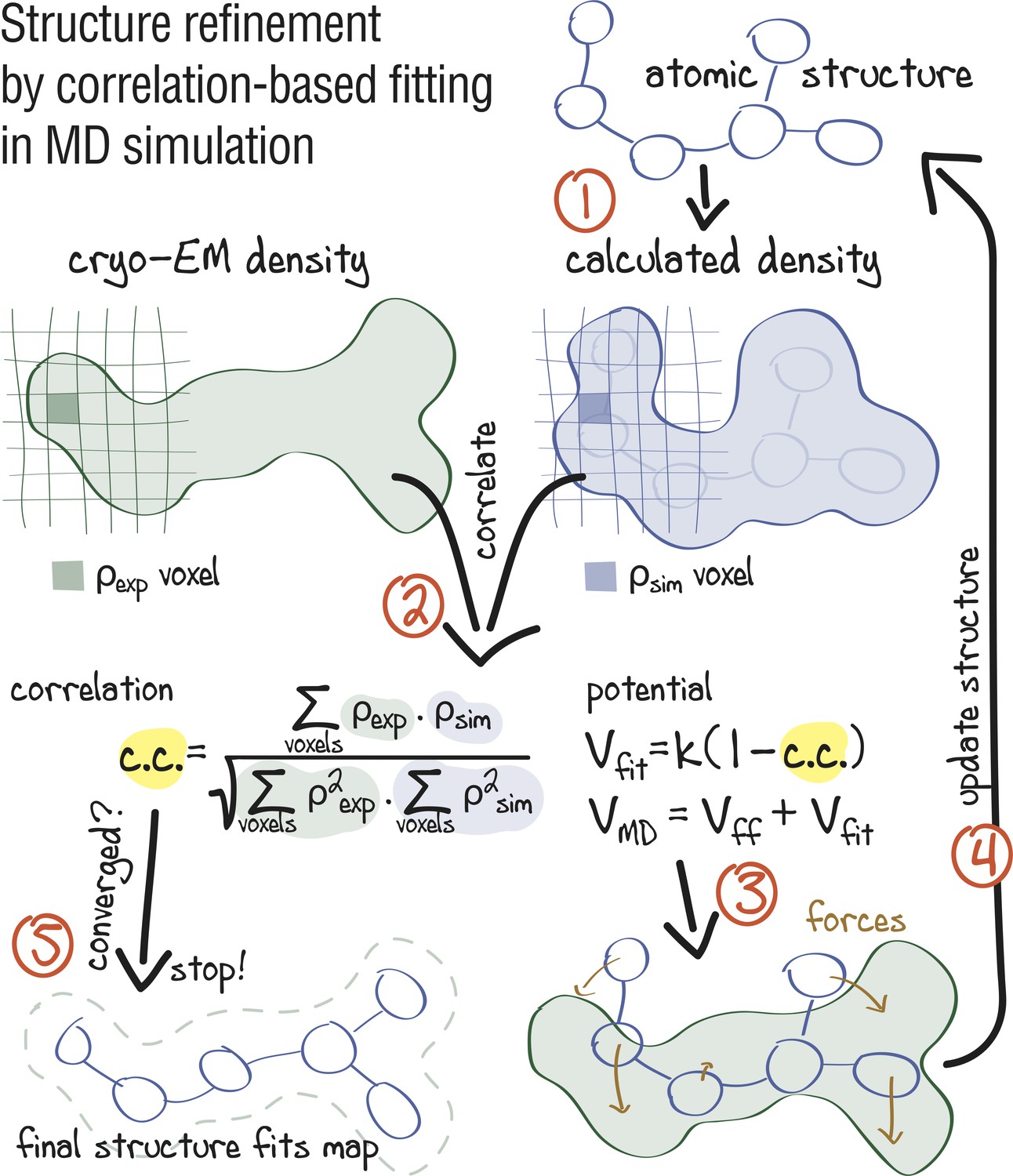
Automated cryo-EM structure refinement using correlation-driven molecular dynamics
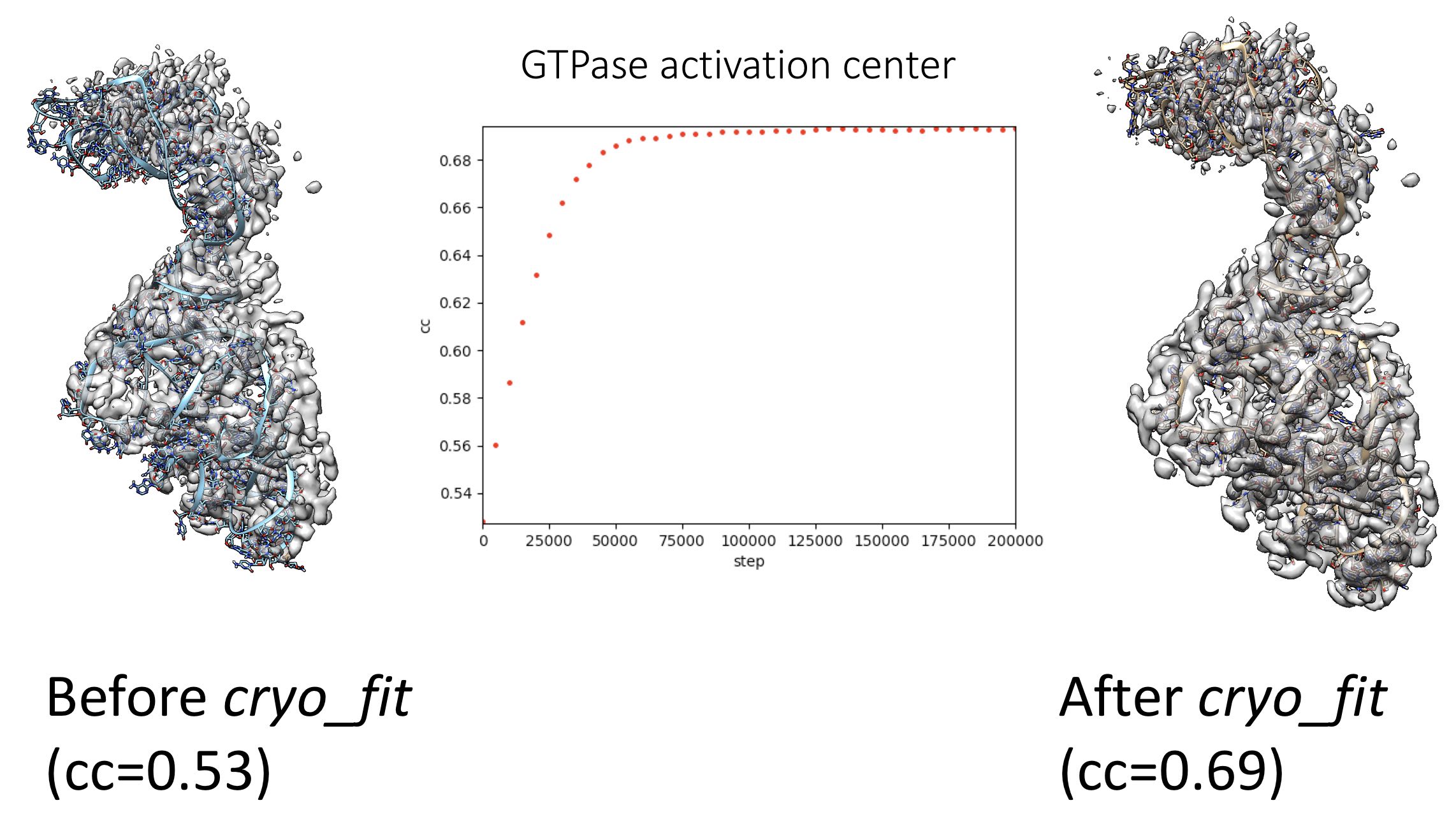
Tutorial: Fit Biomolecules into Cryo-EM Maps using MD Simulation (commandline)

MDSPACE: Extracting Continuous Conformational Landscapes from Cryo-EM Single Particle Datasets Using 3D-to-2D Flexible Fitting based on Molecular Dynamics Simulation - ScienceDirect
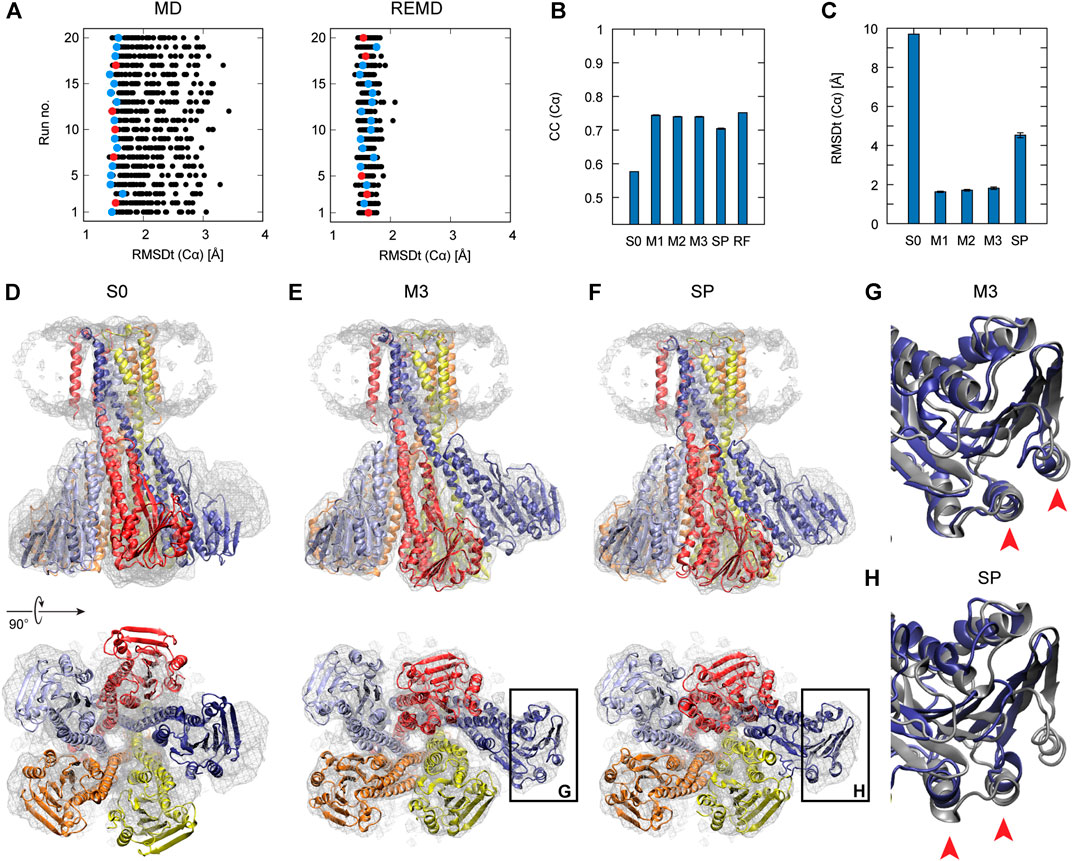
Frontiers Multi-Scale Flexible Fitting of Proteins to Cryo-EM Density Maps at Medium Resolution
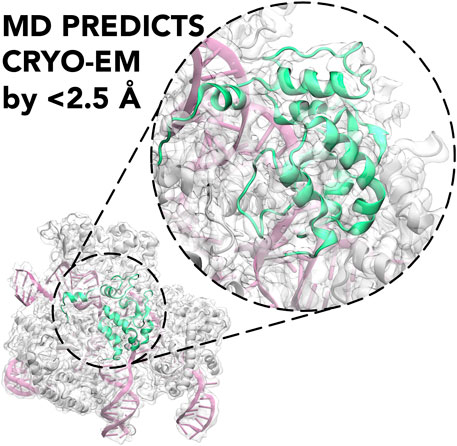
Frontiers Molecular Dynamics to Predict Cryo-EM: Capturing Transitions and Short-Lived Conformational States of Biomolecules







