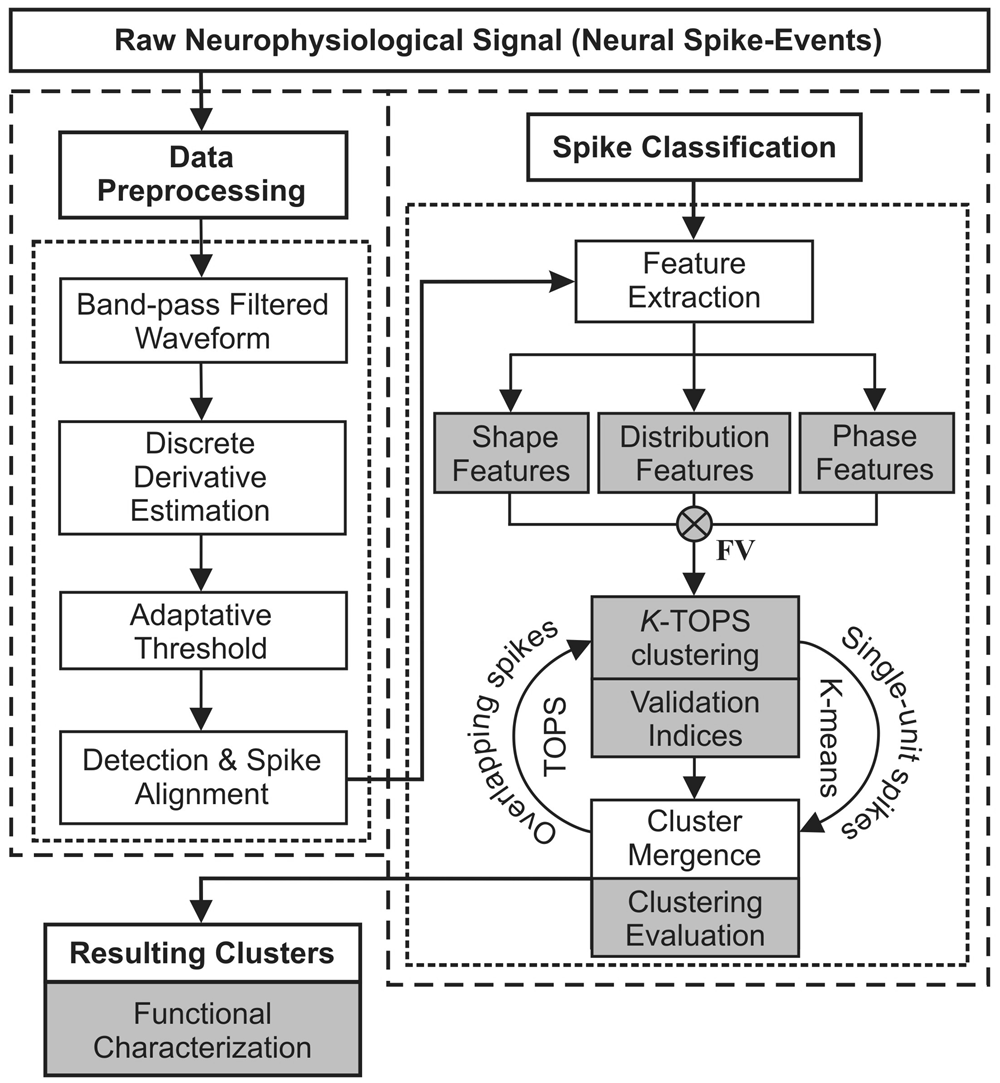
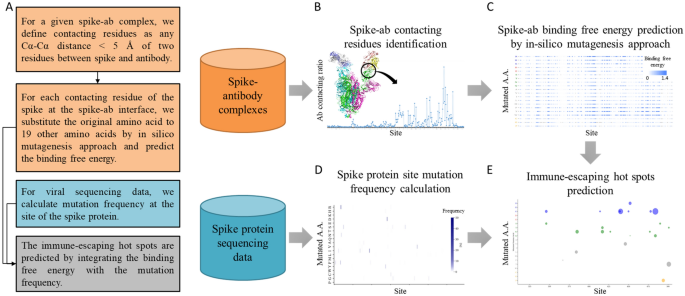
In silico prediction of immune-escaping hot spots for future COVID-19 vaccine design

Antibody responses against SARS-CoV-2 S1 and N proteins in BNT162b2

The precision and recall curve. The threshold estimations of mutation
Effect of SARS-CoV-2 B.1.1.7 mutations on spike protein structure and function

In silico prediction of immune-escaping hot spots for future COVID-19 vaccine design

In silico prediction of immune-escaping hot spots for future COVID-19 vaccine design
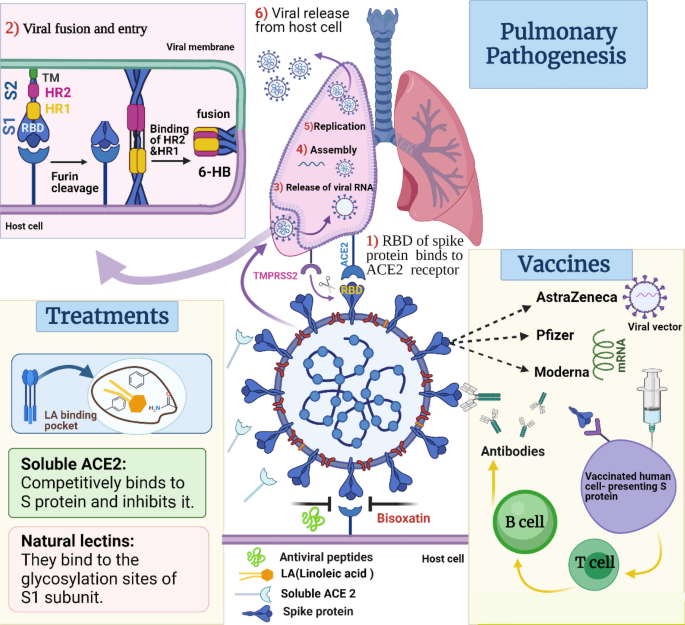
SARS-CoV-2 spike protein: pathogenesis, vaccines, and potential therapies

Full article: Preclinical evaluation of a synthetic peptide vaccine against SARS-CoV-2 inducing multiepitopic and cross-reactive humoral neutralizing and cellular CD4 and CD8 responses
Controlling the SARS-CoV-2 spike glycoprotein conformation

Schematic Diagram of In Silico Vaccine Design Process. (A) Traditional

A comprehensive analysis of the mutational landscape of the newly emerging Omicron (B.1.1.529) variant and comparison of mutations with VOCs and VOIs

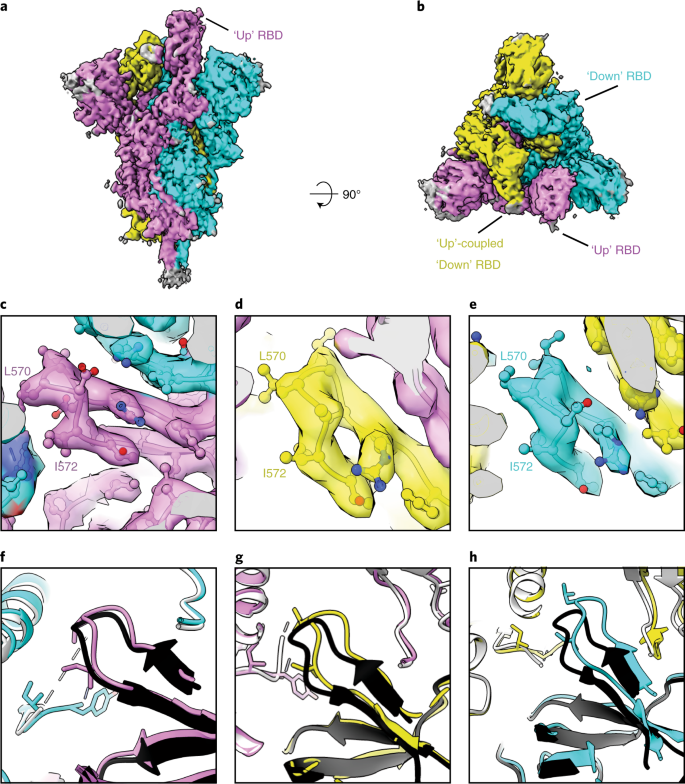
EM structure of the SARS-CoV-2 spike–EY6A Fab complex a,b, Side (a) and
Vaccines April 2023 - Browse Articles

Browse Preprints - Authorea